Reading DICOM images in R
The R packages oro.dicom and oro.nifti [1] provide the ability to import and visualise medical images that have been stored as DICOM [2] or ANALYZE™/NIfTI [3] files. This is handy because DICOM is widely used for clinical data, while NIfTI is a more compact format that stores only 2 files per 3D volume, rather than dozens.
library(oro.dicom)
dcmImages <- readDICOM("/dev/DICOMRT2", verbose = TRUE,
recursive = FALSE, exclude = "sql")
## 412 files to be processed by readDICOM()
##
|
|=================================================================| 100%
A lot of meta-data is associated with each image file, although in the case of de-identified data most of it will be masked out. Nevertheless, the fields that we are interested in for image analysis are present:
dcm.info <- dicomTable(dcmImages$hdr) length(names(dcm.info))
## [1] 137
unique(dcm.info["0008-0008-ImageType"])
## 0008-0008-ImageType
DERIVED PRIMARY AXIAL CT_SOM5 SPI
ORIGINAL PRIMARY AXIAL
<NA>
unique(dcm.info["0018-1110-DistanceSourceToDetector"])
## 0018-1110-DistanceSourceToDetector
1040
1502.15787247627
<NA>
unique(dcm.info["0018-1130-TableHeight"])
## 0018-1130-TableHeight
198
125
<NA>
All of the image data is also available:
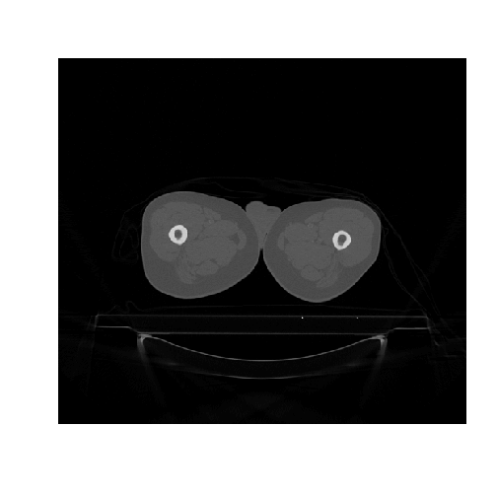
image(t(dcmImages$img[[1]]), col = grey(0:64/64), axes = FALSE,
xlab = "", ylab = "")
A 3D image, such as a CT or MRI, is comprised of multiple DICOM files, one for each slice. Therefore, we need to loop through the files to reconstruct the 3D volume and save it to disk as a NIfTI file:
system.time(for (stack in unique(substring(rownames(dcm.info), 15, 20))) {
if (substring(stack, 1, 2) == "CT") {
print(stack)
index <- which(substring(rownames(dcm.info), 15, 20) == stack)
dcm.stack <- list(hdr = dcmImages$hdr[index], img = dcmImages$img[index])
dcm.nifti <- dicom2nifti(dcm.stack, DIM = 3, descrip = c("Manufacturer",
"ManufacturersModelName"))
print(dcm.nifti)
writeNIfTI(dcm.nifti, paste0("NIfTI/", stack))
}
})
## [1] "CT4693"
## NIfTI-1 format
## Type : nifti
## Data Type : 4 (INT16)
## Bits per Pixel : 16
## Slice Code : 0 (Unknown)
## Intent Code : 0 (None)
## Qform Code : 2 (Aligned_Anat)
## Sform Code : 2 (Aligned_Anat)
## Dimension : 512 x 512 x 86
## Pixel Dimension : 1.27 x 1.27 x 3
## Voxel Units : mm
## Time Units : sec
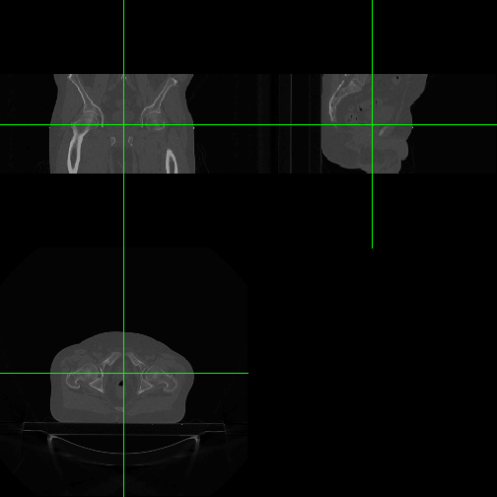
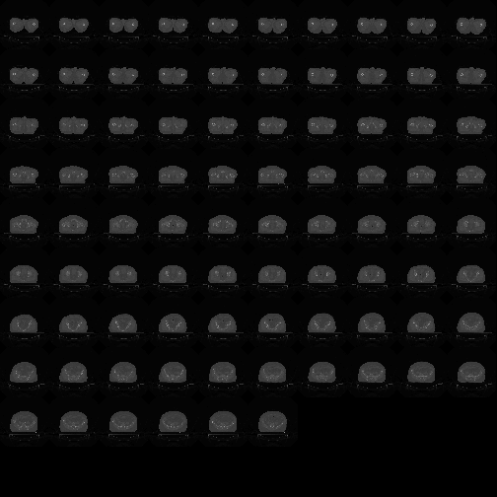
The oro.nifti package also provides some useful visualisations:
orthographic(dcm.nifti, col.crosshairs = "green")
image(dcm.nifti)
References
[1]: B Whitcher, V J Schmid & A Thorton (2011) Working with the DICOM and NIfTI Data Standards in R J Stat Soft 44(6)
[2]: Digital Imaging and Communications in Medicine (DICOM)
[3]: Neuroimaging Informatics Technology Initiative (NIfTI)
If you want to run the above code on the example DICOM-RT file from Jason Dowling’s paper (Inisght Journal, 2013) then you need to make a couple of changes:
dcmImages <- readDICOM("/dev/ITK4SampleDCMRT", verbose = TRUE,recursive = FALSE, exclude = "sql")
dcm.info <- dicomTable(dcmImages$hdr)
system.time(for (stack in unique(substring(rownames(dcm.info), 22, 30))) {
if (substring(stack, 1, 2) == "MR") {
print(stack)
index <- which(substring(rownames(dcm.info), 22, 30) == stack)
dcm.stack <- list(hdr = dcmImages$hdr[index], img = dcmImages$img[index])
dcm.nifti <- dicom2nifti(dcm.stack, DIM = 3, descrip = c("Manufacturer",
"ManufacturersModelName"))
print(dcm.nifti)
writeNIfTI(dcm.nifti, paste0("NIfTI/", stack))
}
})
61 files to be processed by readDICOM()
[1] “MR.1.2.84”
NIfTI-1 format
Type : nifti
Data Type : 4 (INT16)
Bits per Pixel : 16
Slice Code : 0 (Unknown)
Intent Code : 0 (None)
Qform Code : 2 (Aligned_Anat)
Sform Code : 2 (Aligned_Anat)
Dimension : 256 x 256 x 60
Pixel Dimension : 1.48 x 1.48 x 3
Voxel Units : mm
Time Units : sec
user system elapsed
1.74 0.38 2.11